Flatfield calibration#
Inspiration from: https://scripts.iucr.org/cgi-bin/paper?S1600577523001157
Work done for ID31: Scattering of amorphous carbon on a well defined position using a Pilatus CdTe 2M.
There are 9 positions investigated on the detector each of them contains calibration data and a flatfield image. First of all define an object container containing position, calibration, …
[1]:
%matplotlib inline
# Switch from widget <-> inline for documentation purposes
import copy, time, sys
from dataclasses import dataclass
import numpy
import h5py
from matplotlib.pyplot import subplots
import fabio
from silx.resources import ExternalResources
import pyFAI
from pyFAI.gui import jupyter
from pyFAI.gui.jupyter.calib import Calibration
from pyFAI.gui.cli_calibration import AbstractCalibration
print(f"Running pyFAI version {pyFAI.version} on python {sys.version}")
t0 = time.perf_counter()
WARNING:pyFAI.gui.matplotlib:Matplotlib already loaded with backend `inline`, setting its backend to `QtAgg` may not work!
Running pyFAI version 2025.12.0 on python 3.13.1 | packaged by conda-forge | (main, Jan 13 2025, 09:53:10) [GCC 13.3.0]
[2]:
polarization = 0.999
npt = 512
energy = 75 #keV
wavelength = 1e-10*pyFAI.units.hc/energy
detector = pyFAI.detector_factory("Pilatus2M_CdTe")
calibrant = pyFAI.calibrant.CALIBRANT_FACTORY("AgBh")
calibrant.wavelength = wavelength
[3]:
# Here we download the test data
downloader = ExternalResources("flatfield", "http://www.silx.org/pub/pyFAI/testimages")
all_files = downloader.getdir("flatfield_ID31.tar.bz2")
master_file = [i for i in all_files if i.endswith("calibration_0001.h5")][0]
print(master_file)
/tmp/flatfield_testdata_kieffer/flatfield_ID31.tar.bz2__content/flatfield_ID31/calibration_0001.h5
[4]:
@dataclass
class Position:
"""All data related to one of the position"""
position: int
calibration_idx: int
scattering_idx: int
coordinates: tuple=tuple()
calibration_data: object=None
scattering_data: object=None
poni: object=None
ai: object=None
control_points: object=None
flatfield: object=None
@classmethod
def init(cls, h5_file, position, calibration_idx, scattering_idx, detector_name="p3", positioners=("cncx","cncy","cncz")):
with h5py.File(h5_file) as h:
calibration_str = f"{calibration_idx}."
scattering_str = f"{scattering_idx}."
keys = list(h.keys())
ids = [i for i in keys if i.startswith(calibration_str)]
if ids:
entry = h[ids[0]]
calibration_data = entry[f"measurement/{detector_name}"][0]
coordinates = tuple(entry[f"instrument/positioners/{positioner}"][()] for positioner in positioners)
else:
raise IndexError(f"no such Entry {calibration_idx}")
ids = [i for i in keys if i.startswith(scattering_str)]
if ids:
entry = h[ids[0]]
scattering_data = entry[f"measurement/{detector_name}"][0]
coordinates = tuple(entry[f"instrument/positioners/{positioner}"][()] for positioner in positioners)
else:
raise IndexError(f"no such Entry {calibration_idx}")
return cls(position, calibration_idx, scattering_idx, coordinates, calibration_data, scattering_data)
center = Position.init(master_file, "CC", 14, 13)
center
[4]:
Position(position='CC', calibration_idx=14, scattering_idx=13, coordinates=(np.float64(6489.605), np.float64(20.0), np.float64(20.0)), calibration_data=array([[2728, 2784, 2791, ..., 1582, 1636, 1544],
[2664, 2663, 2829, ..., 1542, 1485, 1533],
[2839, 2739, 2674, ..., 1542, 1581, 1478],
...,
[3216, 2998, 3165, ..., 3048, 2992, 3125],
[3121, 3252, 3299, ..., 3086, 3110, 2913],
[3231, 3261, 3414, ..., 3099, 3039, 3020]],
shape=(1679, 1475), dtype=int32), scattering_data=array([[102929, 101856, 105155, ..., 36466, 36234, 35175],
[100320, 98901, 104158, ..., 35047, 34531, 35871],
[102334, 101772, 98380, ..., 35634, 35428, 34703],
...,
[ 96866, 94780, 96978, ..., 95870, 94463, 97045],
[ 97101, 99105, 99604, ..., 97634, 97246, 94603],
[100027, 99620, 102607, ..., 95336, 96377, 94539]],
shape=(1679, 1475), dtype=int32), poni=None, ai=None, control_points=None, flatfield=None)
[5]:
# This contains which scan correspond to what position and if it contains amorphous scattering or calibration data.
data =[None,
Position.init(master_file, 1, 1, 5),
Position.init(master_file, 2, 7, 6),
Position.init(master_file, 3, 8, 9),
Position.init(master_file, 4, 11, 12),
Position.init(master_file, 5, 14, 13),
Position.init(master_file, 6, 15, 16),
Position.init(master_file, 7, 18, 17),
Position.init(master_file, 8, 19, 20),
Position.init(master_file, 9, 22, 21)]
[6]:
#calculate the mask:
mask = -detector.mask.astype(int)
for p in data[1:]:
numpy.minimum(mask, p.scattering_data, out=mask)
numpy.minimum(mask, p.calibration_data, out=mask)
detector.mask = (mask<0).astype(numpy.int8)
fig, ax = subplots()
ax.imshow(detector.mask)
[6]:
<matplotlib.image.AxesImage at 0x7f51004592b0>
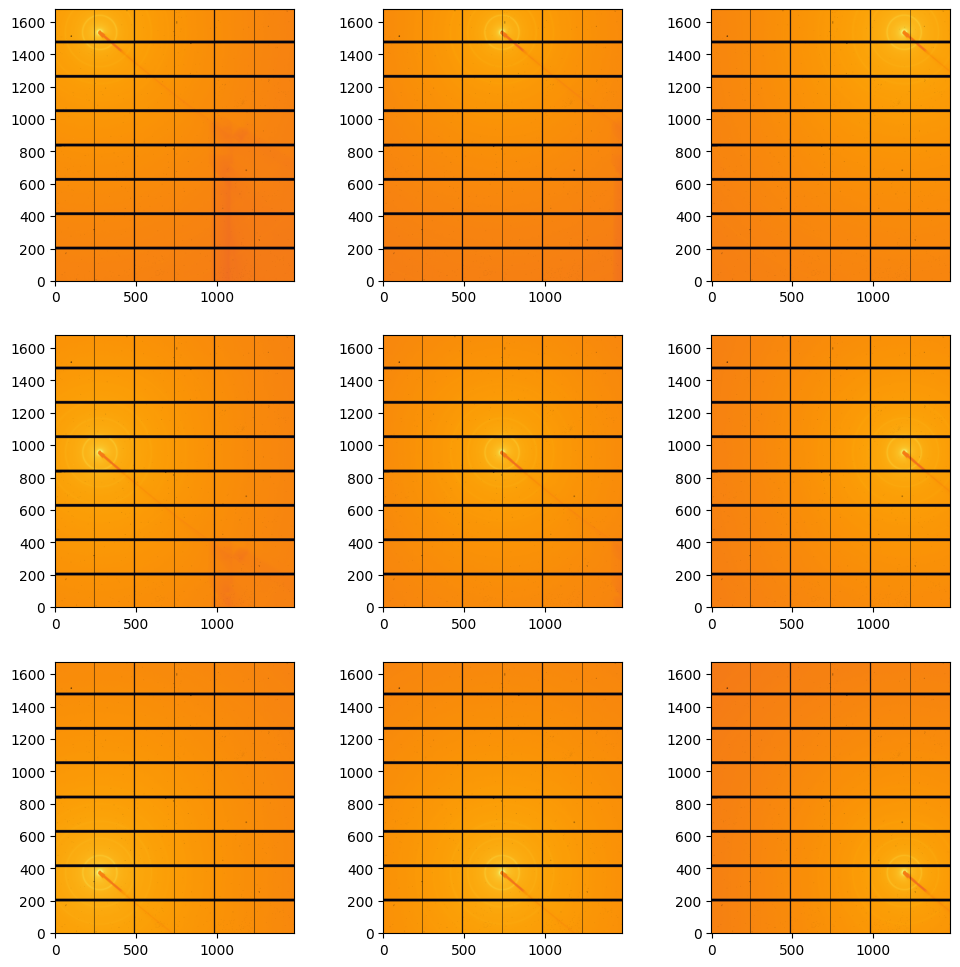
[7]:
#display calibraation scattering:
fig, ax = subplots(3,3, figsize=(12,12))
jupyter.display(data[1].calibration_data, ax=ax[0,2])
jupyter.display(data[2].calibration_data, ax=ax[0,1])
jupyter.display(data[3].calibration_data, ax=ax[0,0])
jupyter.display(data[4].calibration_data, ax=ax[1,0])
jupyter.display(data[5].calibration_data, ax=ax[1,1])
jupyter.display(data[6].calibration_data, ax=ax[1,2])
jupyter.display(data[7].calibration_data, ax=ax[2,2])
jupyter.display(data[8].calibration_data, ax=ax[2,1])
jupyter.display(data[9].calibration_data, ax=ax[2,0]);
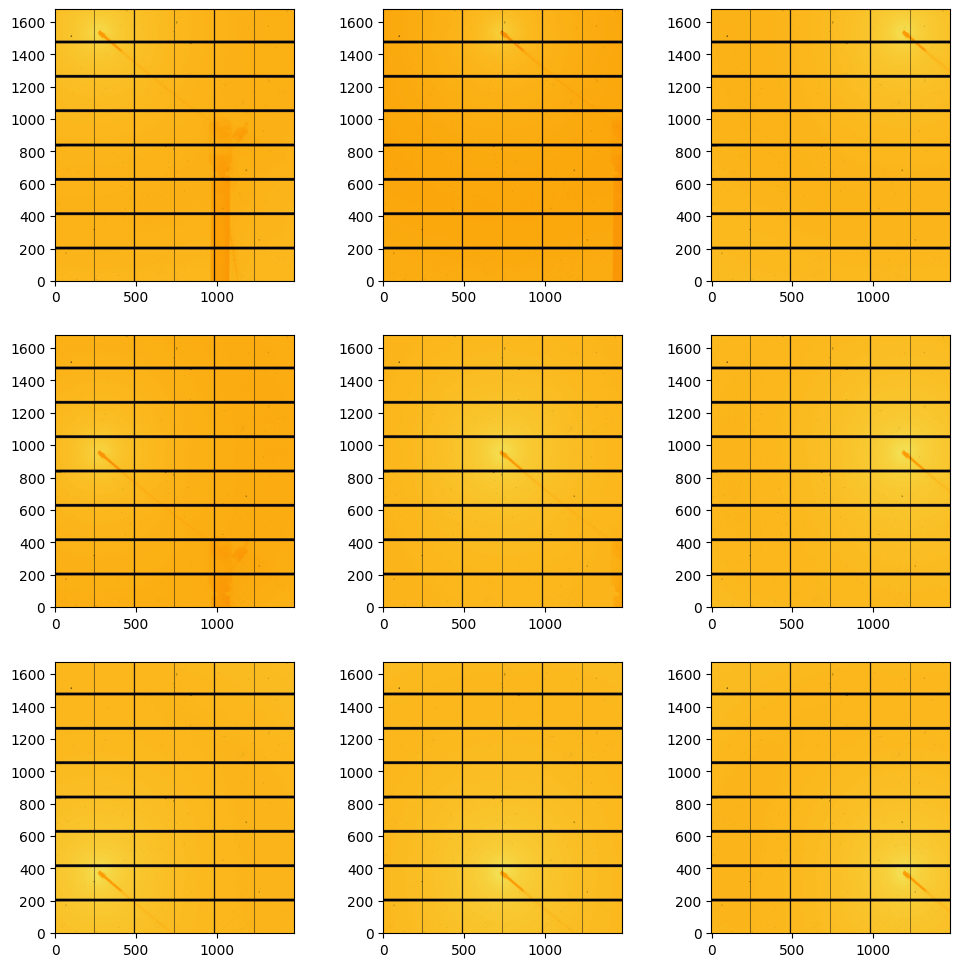
[8]:
#display amorphous scattering:
fig, ax = subplots(3,3, figsize=(12,12))
jupyter.display(data[1].scattering_data, ax=ax[0,2])
jupyter.display(data[2].scattering_data, ax=ax[0,1])
jupyter.display(data[3].scattering_data, ax=ax[0,0])
jupyter.display(data[4].scattering_data, ax=ax[1,0])
jupyter.display(data[5].scattering_data, ax=ax[1,1])
jupyter.display(data[6].scattering_data, ax=ax[1,2])
jupyter.display(data[7].scattering_data, ax=ax[2,2])
jupyter.display(data[8].scattering_data, ax=ax[2,1])
jupyter.display(data[9].scattering_data, ax=ax[2,0]);
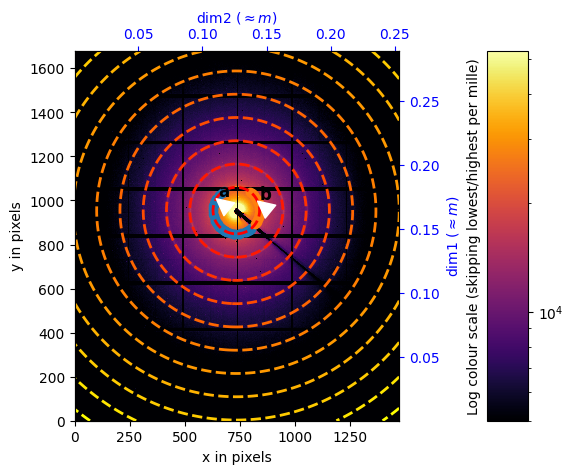
Calibration of the central position#
[9]:
%matplotlib widget
extra_mask = center.calibration_data<5000
# switch to widget mode ... for calibration purpose. Use right click.
calib = Calibration(center.calibration_data,
calibrant=calibrant,
wavelength=calibrant.wavelength,
detector=copy.deepcopy(detector),
mask=extra_mask) # Mind the mask option mangles the detector's mask !
[11]:
input("Please perform the calibration in the previous cell before going on ... use the right-click")
%matplotlib inline
fig, ax = subplots()
Please perform the calibration in the previous cell before going on ... use the right-click
[14]:
#Reset the mask
calib.mask=None
calib.geoRef.detector = detector
print(calib.geoRef)
f2d = calib.geoRef.getFit2D()
f2d["tilt"] = 0
# f2d.pop("splineFile")
print(f2d)
calib.geoRef.setFit2D(**f2d)
print(calib.geoRef)
calib.fixed += ["rot1", "rot2"]
print(calib.fixed)
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.397882e+00 m PONI= 1.403805e-01, 9.224468e-02 m rot1=-0.005337 rot2=0.003714 rot3=0.000000 rad
DirectBeamDist= 6398.017 mm Center: x=734.835, y=954.302 pix Tilt= 0.373° tiltPlanRotation= 34.830° λ= 0.165Å
DirectBeamDist= 6398.017 mm Center: x=734.835, y=954.302 pix Tilt= 0.000° tiltPlanRotation= 34.830° λ= 0.165Å
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.398017e+00 m PONI= 1.641400e-01, 1.263916e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6398.017 mm Center: x=734.835, y=954.302 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
Fixed parameters: wavelength, rot1, rot3, rot2.
[15]:
logger = pyFAI.gui.cli_calibration.logger
from silx.image import marchingsquares
def extract_cpt(self, method="massif", pts_per_deg=1.0, max_rings=numpy.iinfo(int).max):
"""
Performs an automatic keypoint extraction:
Can be used in recalib or in calib after a first calibration has been performed.
:param method: method for keypoint extraction
:param pts_per_deg: number of control points per azimuthal degree (increase for better precision)
:param max_rings: extract at most max_rings
"""
logger.info("in extract_cpt with method %s", method)
assert self.ai
assert self.calibrant
assert self.peakPicker
self.peakPicker.reset()
self.peakPicker.init(method, False)
if self.geoRef:
self.ai.set_config(self.geoRef.get_config())
tth = numpy.array([i for i in self.calibrant.get_2th() if i is not None])
tth = numpy.unique(tth)
tth_min = numpy.zeros_like(tth)
tth_max = numpy.zeros_like(tth)
delta = (tth[1:] - tth[:-1]) / 4.0
tth_max[:-1] = delta
tth_max[-1] = delta[-1]
tth_min[1:] = -delta
tth_min[0] = -delta[0]
tth_max += tth
tth_min += tth
shape = self.peakPicker.data.shape
if self.geoRef:
ttha = self.geoRef.center_array(shape, unit="2th_rad", scale=False)
chia = self.geoRef.center_array(shape, unit="chi_rad", scale=False)
else:
ttha = self.ai.center_array(shape, unit="2th_rad", scale=False)
chia = self.ai.center_array(shape, unit="chi_rad", scale=False)
rings = 0
self.peakPicker.sync_init()
if self.max_rings is None:
self.max_rings = tth.size
ms = marchingsquares.MarchingSquaresMergeImpl(ttha, self.mask, use_minmax_cache=True)
for i in range(tth.size):
if rings >= min(self.max_rings, max_rings):
break
mask1 = numpy.logical_and(ttha >= tth_min[i], ttha < tth_max[i])
if self.mask is not None:
numpy.logical_and(mask1, numpy.logical_not(self.mask), out=mask1)
size = mask1.sum(dtype=int)
if (size > 0):
rings += 1
self.peakPicker.massif_contour(mask1)
# if self.gui:
# self.peakPicker.widget.update()
sub_data = self.peakPicker.data.ravel()[numpy.where(mask1.ravel())]
mean = sub_data.mean(dtype=numpy.float64)
std = sub_data.std(dtype=numpy.float64)
upper_limit = mean + std
mask2 = numpy.logical_and(self.peakPicker.data > upper_limit, mask1)
size2 = mask2.sum(dtype=int)
if size2 < 1000:
upper_limit = mean
numpy.logical_and(self.peakPicker.data > upper_limit, mask1, out=mask2)
size2 = mask2.sum()
# length of the arc:
points = ms.find_pixels(tth[i])
seeds = set((i[0], i[1]) for i in points if mask2[i[0], i[1]])
# max number of points: 360 points for a full circle
azimuthal = chia[points[:, 0].clip(0, self.peakPicker.data.shape[0]), points[:, 1].clip(0, self.peakPicker.data.shape[1])]
nb_deg_azim = numpy.unique(numpy.rad2deg(azimuthal).round()).size
keep = int(nb_deg_azim * pts_per_deg)
if keep == 0:
continue
dist_min = len(seeds) / 2.0 / keep
# why 3.0, why not ?
logger.info("Extracting datapoint for ring %s (2theta = %.2f deg); "
"searching for %i pts out of %i with I>%.1f, dmin=%.1f" %
(i, numpy.degrees(tth[i]), keep, size2, upper_limit, dist_min))
_res = self.peakPicker.peaks_from_area(mask=mask2, Imin=upper_limit, keep=keep, method=method, ring=i, dmin=dist_min, seed=seeds)
if self.basename:
self.peakPicker.points.save(self.basename + ".npt")
if self.weighted:
self.data = self.peakPicker.points.getWeightedList(self.peakPicker.data)
else:
self.data = self.peakPicker.points.getList()
if self.geoRef:
self.geoRef.data = numpy.array(self.data, dtype=numpy.float64)
Calibration.extract_cpt = extract_cpt
[16]:
calib.extract_cpt(max_rings=4)
[17]:
calib.refine()
Before refinement, the geometry is:
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.398017e+00 m PONI= 1.641400e-01, 1.263916e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6398.017 mm Center: x=734.835, y=954.302 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.411722e+00 m PONI= 1.641389e-01, 1.264064e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6411.722 mm Center: x=734.921, y=954.296 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.411722e+00 m PONI= 1.641389e-01, 1.264064e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6411.722 mm Center: x=734.921, y=954.296 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
[18]:
ai = calib.geoRef.promote("pyFAI.integrator.azimuthal.AzimuthalIntegrator")
it = ai.integrate1d(center.scattering_data, npt, polarization_factor=polarization, error_model="no", method=("no", "csr", "cython"))
sc = ai.sigma_clip(center.scattering_data, npt, polarization_factor=polarization, error_model="azimuthal", method=("no", "csr", "cython"),
thres=0, max_iter=3)
md = ai.medfilt1d_ng(center.scattering_data, npt, polarization_factor=polarization, method=("full", "csr", "cython"))
[19]:
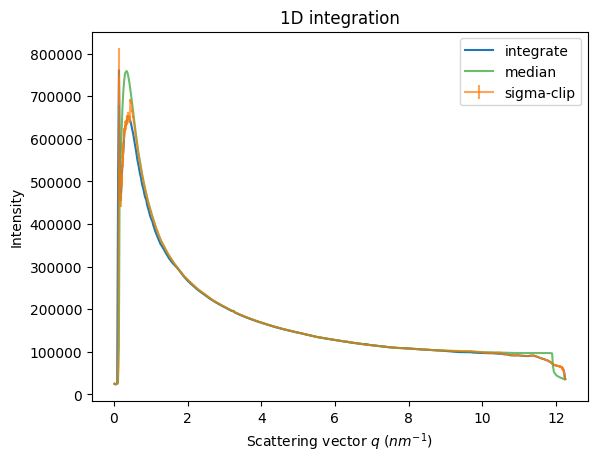
fig, ax = subplots()
ax = jupyter.plot1d(it, label="integrate", ax=ax)
ax.errorbar(*sc, alpha=0.7, label="sigma-clip")
ax.plot(*md, alpha=0.7, label="median")
# ax.set_yscale("log")
ax.legend();
[20]:
# Approximate polarization correction needed:
print(f"Approximate polarization factor: {ai.guess_polarization(center.scattering_data, unit='q_nm^-1', target_rad=10):.4f}")
Approximate polarization factor: 0.9985
[21]:
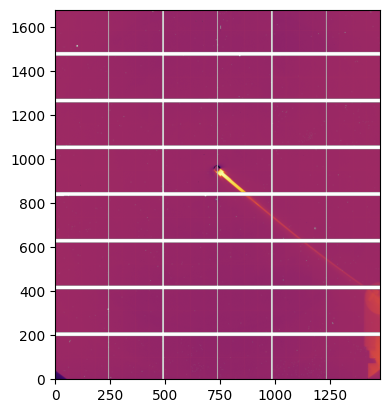
# median filter provides the smoothest curve achievable
rebuilt = ai.calcfrom1d(md.radial,
md.intensity,
detector.shape,
dim1_unit=pyFAI.units.Q_NM,
polarization_factor=polarization)
flat = rebuilt/center.scattering_data
flat[numpy.where(detector.mask)] = numpy.nan
flat[center.scattering_data<=0] = numpy.nan
jupyter.display(flat);
/tmp/ipykernel_209755/625551917.py:7: RuntimeWarning: divide by zero encountered in divide
flat = rebuilt/center.scattering_data
Calculate the approximate correction of the other positions#
[22]:
dx,dy,dz = numpy.array(data[1].coordinates)-center.coordinates
ai1 = copy.copy(ai)
ai1.poni1 += dz*0.001
ai1.poni2 += dy*0.001
ai1
[22]:
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.411722e+00 m PONI= 2.641389e-01, 2.051464e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6411.722 mm Center: x=1192.712, y=1535.691 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
[23]:
AbstractCalibration.extract_cpt = extract_cpt
[24]:
calib1 = AbstractCalibration(data[1].calibration_data, detector.mask, detector, wavelength=wavelength, calibrant=calibrant)
calib1.preprocess()
calib1.data = []
calib1.geoRef = calib1.initgeoRef()
calib1.geoRef.set_config(ai1.get_config())
[24]:
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.411722e+00 m PONI= 2.641389e-01, 2.051464e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6411.722 mm Center: x=1192.712, y=1535.691 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
[25]:
calib1.extract_cpt(max_rings=4)
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
[26]:
calib1.fixed += ["rot1", "rot2"]
calib1.geoRef.refine3(fix=calib1.fixed)
calib1.fixed
[26]:
Fixed parameters: wavelength, rot1, rot3, rot2.
[27]:
calib1.geoRef
[27]:
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.429301e+00 m PONI= 2.640936e-01, 2.050724e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6429.301 mm Center: x=1192.281, y=1535.428 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
[28]:
data[1].ai = pyFAI.load(calib1.geoRef)
data[1].control_points = calib1.peakPicker.points
data[5].ai = pyFAI.load(calib.geoRef)
data[5].control_points = calib.peakPicker.points
Perform the geometry extraction for each of the position:#
[29]:
for idx in [2,3,4,6,7,8,9]:
dx,dy,dz = numpy.array(data[idx].coordinates)-center.coordinates
ain = copy.copy(ai)
ain.poni1 += dz*0.001
ain.poni2 += dy*0.001
calibn = AbstractCalibration(data[idx].calibration_data, detector.mask, detector, wavelength=wavelength, calibrant=calibrant)
calibn.preprocess()
calibn.data = []
calibn.geoRef = calib1.initgeoRef()
calibn.geoRef.set_config(ain.get_config())
calibn.extract_cpt(max_rings=4)
calibn.fixed += ["rot1", "rot2"]
calibn.geoRef.refine3(fix=calibn.fixed)
print(f"#### Position {idx} ####")
print(calibn.geoRef)
data[idx].ai = pyFAI.load(calibn.geoRef)
data[idx].control_points = calibn.peakPicker.points
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
#### Position 2 ####
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.431163e+00 m PONI= 2.641045e-01, 1.262797e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6431.163 mm Center: x=734.184, y=1535.491 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
#### Position 3 ####
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.429291e+00 m PONI= 2.640419e-01, 4.748285e-02 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6429.291 mm Center: x=276.063, y=1535.127 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
#### Position 4 ####
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.429678e+00 m PONI= 1.641200e-01, 4.762339e-02 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6429.678 mm Center: x=276.880, y=954.186 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
#### Position 6 ####
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.429827e+00 m PONI= 1.641299e-01, 2.051724e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6429.827 mm Center: x=1192.863, y=954.244 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
#### Position 7 ####
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.431333e+00 m PONI= 6.420481e-02, 2.052253e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6431.333 mm Center: x=1193.171, y=373.284 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
#### Position 8 ####
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.432639e+00 m PONI= 6.413222e-02, 1.264375e-01 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6432.639 mm Center: x=735.102, y=372.862 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
ERROR:root:No diffraction image available => not showing the contour
#### Position 9 ####
Detector Pilatus CdTe 2M PixelSize= 172µm, 172µm BottomRight (3)
Wavelength= 0.165312 Å
SampleDetDist= 6.431421e+00 m PONI= 6.412127e-02, 4.770164e-02 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 6431.421 mm Center: x=277.335, y=372.798 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 0.165Å
[30]:
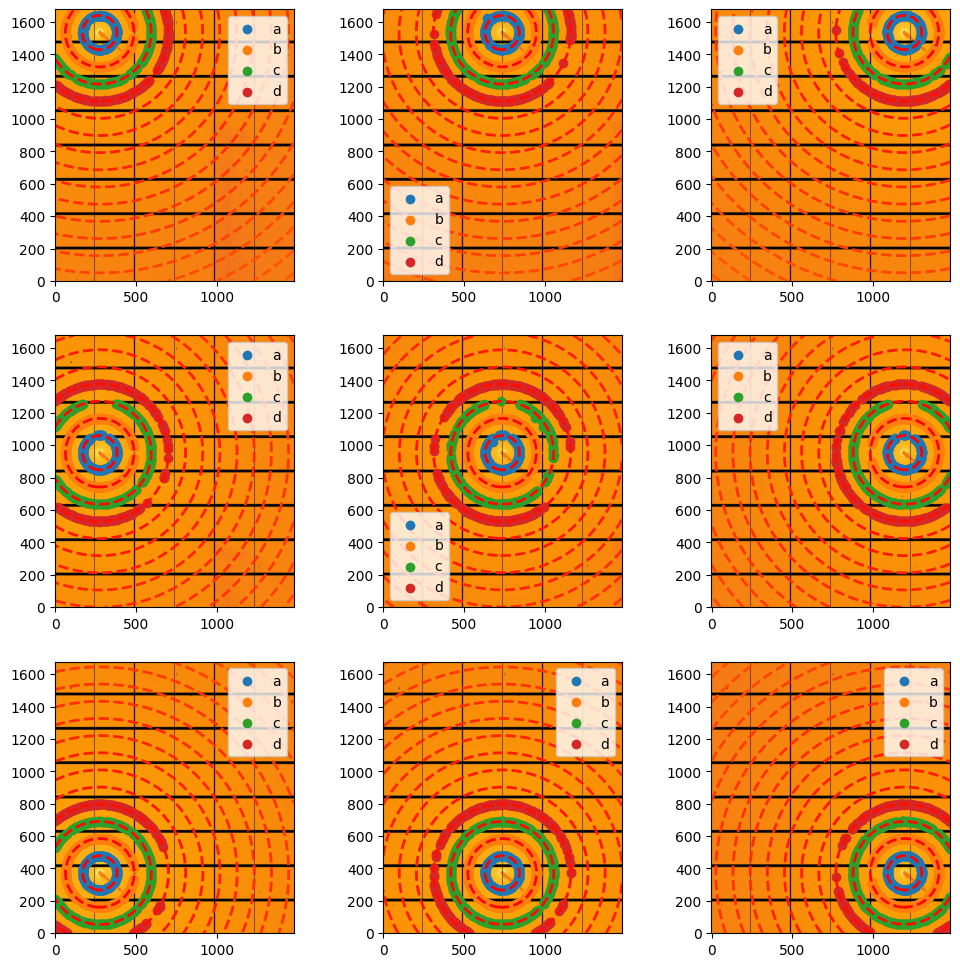
#display scattering:
fig, ax = subplots(3,3, figsize=(12,12))
jupyter.display(data[1].calibration_data, ai=data[1].ai, cp=data[1].control_points, ax=ax[0,2])
jupyter.display(data[2].calibration_data, ai=data[2].ai, cp=data[2].control_points, ax=ax[0,1])
jupyter.display(data[3].calibration_data, ai=data[3].ai, cp=data[3].control_points, ax=ax[0,0])
jupyter.display(data[4].calibration_data, ai=data[4].ai, cp=data[4].control_points, ax=ax[1,0])
jupyter.display(data[5].calibration_data, ai=data[5].ai, cp=data[5].control_points, ax=ax[1,1])
jupyter.display(data[6].calibration_data, ai=data[6].ai, cp=data[6].control_points, ax=ax[1,2])
jupyter.display(data[7].calibration_data, ai=data[7].ai, cp=data[7].control_points, ax=ax[2,2])
jupyter.display(data[8].calibration_data, ai=data[8].ai, cp=data[8].control_points, ax=ax[2,1])
jupyter.display(data[9].calibration_data, ai=data[9].ai, cp=data[9].control_points, ax=ax[2,0]);
Extract the flatfield for all positions#
[31]:
for p in data[1:]:
md = p.ai.medfilt1d_ng(p.scattering_data, npt, polarization_factor=polarization, method=("full", "csr", "cython"))
rebuilt = p.ai.calcfrom1d(md.radial, md.intensity, detector.shape, dim1_unit=pyFAI.units.Q_NM, polarization_factor=polarization)
flat = rebuilt / p.scattering_data
flat[numpy.where(detector.mask)] = numpy.nan
flat[p.scattering_data<=0] = numpy.nan
p.flat = flat
/tmp/ipykernel_209755/3685900361.py:4: RuntimeWarning: divide by zero encountered in divide
flat = rebuilt / p.scattering_data
[32]:
#display flat:
fig, ax = subplots(3,3, figsize=(12,12))
jupyter.display(data[1].flat, ax=ax[0,2])
jupyter.display(data[2].flat, ax=ax[0,1])
jupyter.display(data[3].flat, ax=ax[0,0])
jupyter.display(data[4].flat, ax=ax[1,0])
jupyter.display(data[5].flat, ax=ax[1,1])
jupyter.display(data[6].flat, ax=ax[1,2])
jupyter.display(data[7].flat, ax=ax[2,2])
jupyter.display(data[8].flat, ax=ax[2,1])
jupyter.display(data[9].flat, ax=ax[2,0]);
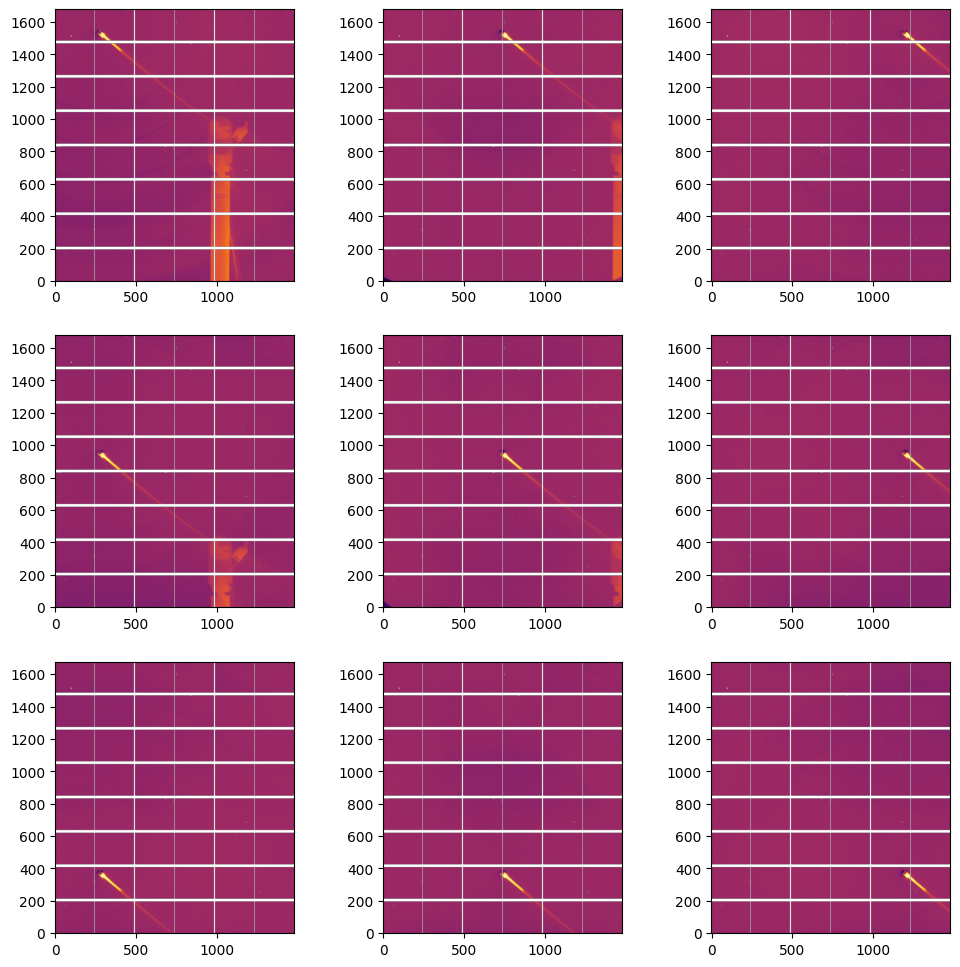
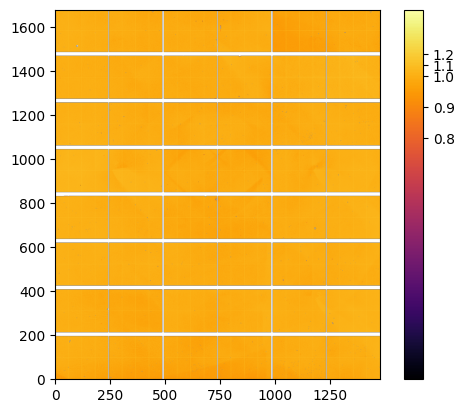
The final Flatfield is the median of the flats calculated on the 9 positions#
[33]:
flat_stack = numpy.array([p.flat for p in data[1:]])
flat = numpy.nanmedian(flat_stack, axis=0)
/tmp/ipykernel_209755/4120576390.py:2: RuntimeWarning: All-NaN slice encountered
flat = numpy.nanmedian(flat_stack, axis=0)
[34]:
ax = jupyter.display(flat)
cb = ax.figure.colorbar(ax.images[0]);
pos = numpy.linspace(0.8,1.2, 5)
ticks = [str(i) for i in pos]
cb.set_ticks(pos, labels=ticks);
[35]:
fabio.edfimage.EdfImage(data=flat.astype("float32")).write("flat.edf")
[36]:
print(f"Total run time: {time.perf_counter()-t0:.3f}s.")
Total run time: 143.419s.