2D Integration in non-azimuthal space#
In this tutorial, an image is intgrated in qx/qy space instead of 2theta/chi. More fancy spaces are even possible …
[1]:
%matplotlib inline
# %matplotlib widget
import os
os.environ["PYOPENCL_COMPILER_OUTPUT"] = "0"
[2]:
import numpy
import pyFAI
from pyFAI.test.utilstest import UtilsTest
import fabio
from pyFAI.gui import jupyter
from pyFAI import units
from pyFAI.method_registry import Method,IntegrationMethod
print("pyFAI version:", pyFAI.version)
pyFAI version: 2025.12.0
[3]:
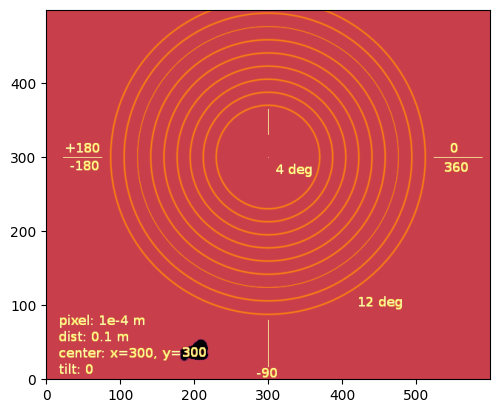
img = fabio.open(UtilsTest.getimage("moke.tif")).data
jupyter.display(img);
[4]:
# According to the description in the image, we have:
det = pyFAI.detector_factory("Detector", {"pixel1":1e-4,"pixel2":1e-4})
ai = pyFAI.load({"detector": det, "wavelength": 1e-10})
ai.setFit2D(100, 300, 300)
ai
[4]:
Detector Detector PixelSize= 100µm, 100µm BottomRight (3)
Wavelength= 1.000000 Å
SampleDetDist= 1.000000e-01 m PONI= 3.000000e-02, 3.000000e-02 m rot1=0.000000 rot2=0.000000 rot3=0.000000 rad
DirectBeamDist= 100.000 mm Center: x=300.000, y=300.000 pix Tilt= 0.000° tiltPlanRotation= 0.000° λ= 1.000Å
[5]:
len(IntegrationMethod.select_method(dim=2))
[5]:
48
[6]:
for m in IntegrationMethod.select_method(dim=2):
print(m, end="")
if m.algorithm=="histogram" and m.target is not None:
print("OpenCL histogram skipped")
continue
try:
res = ai.integrate2d(img, 400,400, method=m, unit=("qx_nm^-1","qy_nm^-1"))
except:
print("broken")
else:
print(f"{res[1].min():.2f}<qx<{res[1].max():.2f}, {res[2].min():.2f}<qy<{res[2].max():.2f} <I>={numpy.nanmean(res[0]):.2f} ± {numpy.nanstd(res[0]):.2f}")
WARNING:pyFAI.geometry.core:No fast path for space: `qx`
WARNING:pyFAI.geometry.core:No fast path for space: `qy`
IntegrationMethod(2d int, no split, histogram, python)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, no split, histogram, cython)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, histogram, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, histogram, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, pseudo split, histogram, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=2.44 ± 13.72
IntegrationMethod(2d int, no split, CSR, cython)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, CSR, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, CSR, python)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, CSR, python)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, CSC, cython)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, CSC, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, CSC, python)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, CSC, python)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, bbox split, LUT, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, LUT, cython)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, full split, LUT, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, CSR, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, CSR, python)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, CSC, cython)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, CSC, python)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, histogram, OpenCL, NVIDIA CUDA / NVIDIA RTX A5000)OpenCL histogram skipped
IntegrationMethod(2d int, no split, histogram, OpenCL, NVIDIA CUDA / Quadro P2200)OpenCL histogram skipped
IntegrationMethod(2d int, no split, histogram, OpenCL, Portable Computing Language / cpu-haswell-AMD Ryzen Threadripper PRO 3975WX 32-Cores)OpenCL histogram skipped
IntegrationMethod(2d int, no split, histogram, OpenCL, Intel(R) OpenCL / AMD Ryzen Threadripper PRO 3975WX 32-Cores)OpenCL histogram skipped
IntegrationMethod(2d int, bbox split, CSR, OpenCL, NVIDIA CUDA / NVIDIA RTX A5000)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, CSR, OpenCL, NVIDIA CUDA / NVIDIA RTX A5000)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, CSR, OpenCL, NVIDIA CUDA / Quadro P2200)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, CSR, OpenCL, NVIDIA CUDA / Quadro P2200)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, CSR, OpenCL, Portable Computing Language / cpu-haswell-AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, CSR, OpenCL, Portable Computing Language / cpu-haswell-AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, CSR, OpenCL, Intel(R) OpenCL / AMD Ryzen Threadripper PRO 3975WX 32-Cores)
/users/kieffer/.venv/py313/lib/python3.13/site-packages/pyopencl/cache.py:496: CompilerWarning: Non-empty compiler output encountered. Set the environment variable PYOPENCL_COMPILER_OUTPUT=1 to see more.
_create_built_program_from_source_cached(
/users/kieffer/.venv/py313/lib/python3.13/site-packages/pyopencl/cache.py:500: CompilerWarning: Non-empty compiler output encountered. Set the environment variable PYOPENCL_COMPILER_OUTPUT=1 to see more.
prg.build(options_bytes, devices)
-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, CSR, OpenCL, Intel(R) OpenCL / AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, full split, CSR, OpenCL, NVIDIA CUDA / NVIDIA RTX A5000)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, CSR, OpenCL, NVIDIA CUDA / Quadro P2200)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, CSR, OpenCL, Portable Computing Language / cpu-haswell-AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, CSR, OpenCL, Intel(R) OpenCL / AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, bbox split, LUT, OpenCL, NVIDIA CUDA / NVIDIA RTX A5000)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, LUT, OpenCL, NVIDIA CUDA / NVIDIA RTX A5000)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, LUT, OpenCL, NVIDIA CUDA / Quadro P2200)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, LUT, OpenCL, NVIDIA CUDA / Quadro P2200)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, LUT, OpenCL, Portable Computing Language / cpu-haswell-AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, LUT, OpenCL, Portable Computing Language / cpu-haswell-AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, bbox split, LUT, OpenCL, Intel(R) OpenCL / AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, no split, LUT, OpenCL, Intel(R) OpenCL / AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.17<qx<18.17, -18.18<qy<12.31 <I>=10.60 ± 26.61
IntegrationMethod(2d int, full split, LUT, OpenCL, NVIDIA CUDA / NVIDIA RTX A5000)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, LUT, OpenCL, NVIDIA CUDA / Quadro P2200)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, LUT, OpenCL, Portable Computing Language / cpu-haswell-AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
IntegrationMethod(2d int, full split, LUT, OpenCL, Intel(R) OpenCL / AMD Ryzen Threadripper PRO 3975WX 32-Cores)-18.20<qx<18.20, -18.21<qy<12.34 <I>=10.55 ± 24.38
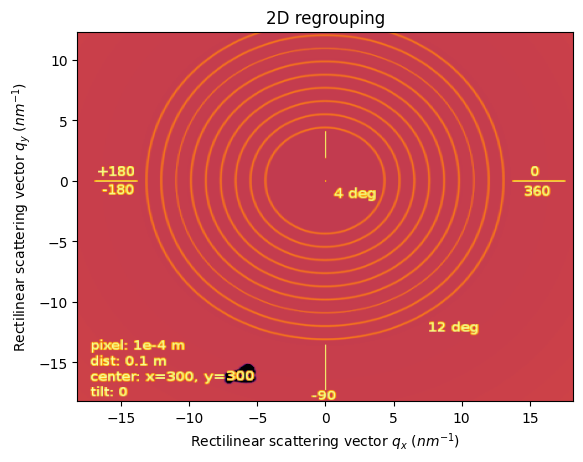
[7]:
m = IntegrationMethod.select_method(dim=2, split="full", algo="CSR")[0]
print(m)
ai.reset()
res = ai.integrate2d(img, 400,400, method=m, unit=("qx_nm^-1", "qy_nm^-1"))
jupyter.plot2d(res);
IntegrationMethod(2d int, full split, CSR, cython)
WARNING:pyFAI.geometry.core:No fast path for space: `qx`
WARNING:pyFAI.geometry.core:No fast path for space: `qy`
[8]:
ai._cached_array.keys()
[8]:
dict_keys(['cos_incidence', 'solid_angle#3.0_crc', 'solid_angle#3.0', 'qx_corner', 'qy_corner'])
[9]:
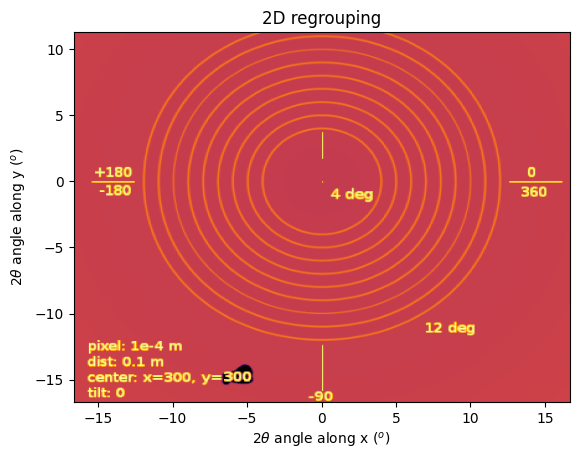
# Now with a new space: 2thx, 2thy:
tthx = pyFAI.units.register_radial_unit("tthx_deg",
scale=180.0/numpy.pi,
label=r"$2\theta$ angle along x ($^{o}$)",
formula="arctan2(x,z)",
short_name="2thx",
unit_symbol="^{o}",
positive=False)
tthy = pyFAI.units.register_azimuthal_unit("tthy_deg",
scale=180.0/numpy.pi,
label=r"$2\theta$ angle along y ($^{o}$)",
formula="arctan2(y,z)",
short_name="2thy",
unit_symbol="^{o}",
positive=False)
res = ai.integrate2d(img, 400,400, method=m, unit=("tthx_deg", "tthy_deg"))
jupyter.plot2d(res);
WARNING:pyFAI.geometry.core:No fast path for space: `tthx`
WARNING:pyFAI.geometry.core:No fast path for space: `tthy`
[10]:
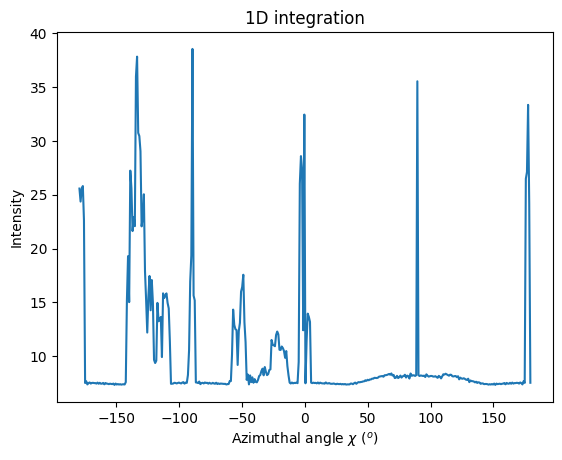
#Nota: it is also possible to integrate along the radial dim ... but for now, no way to provide limitation in radius.
jupyter.plot1d(ai.integrate1d(img, 400, unit="chi_deg", method=("no", "histogram", "cython")));
[ ]:
[ ]: