Sample Code¶
All sample codes can be downloaded as a zip file: silx_examples.zip.
silx.gui sample code¶
| Source | Screenshot | Description |
|---|---|---|
animatedicons.py |
Display available project icons using Qt. | |
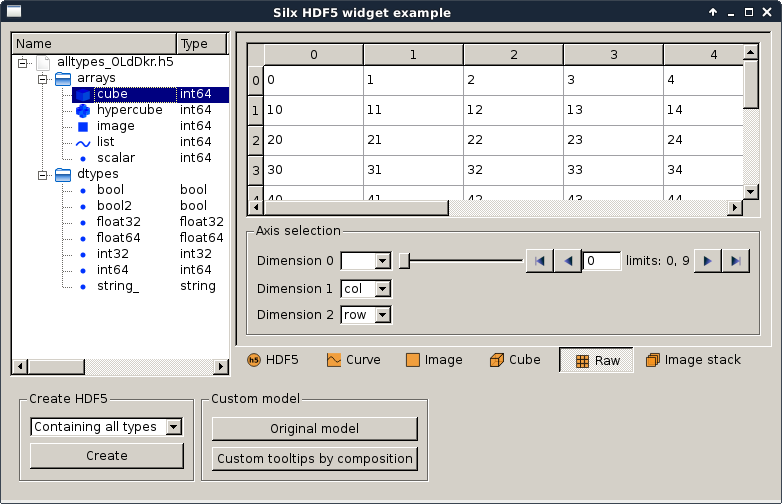
customHdf5TreeModel.py |
|
Qt Hdf5 widget examples |
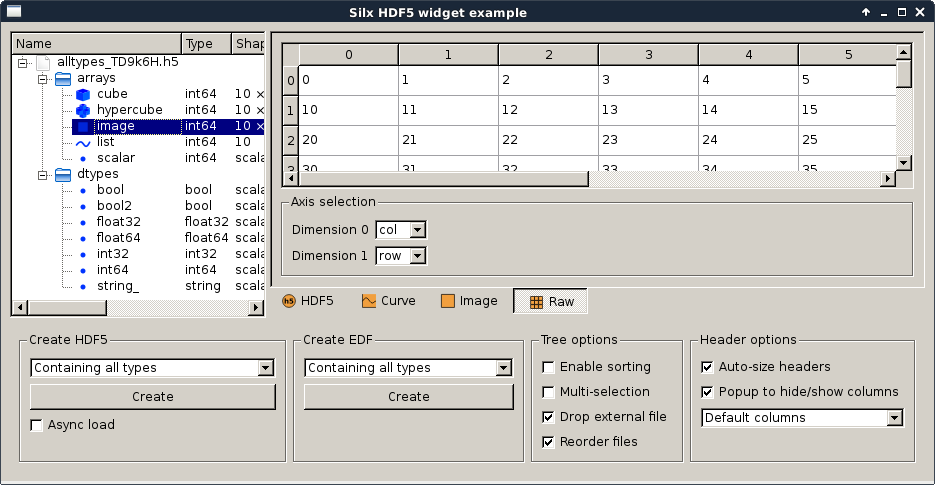
hdf5widget.py |
|
Qt Hdf5 widget examples Note This module has a dependency on the h5py library, which is not a mandatory dependency for silx. You might need to install it if you don’t already have it. |
icons.py |
Display available project icons using Qt. | |
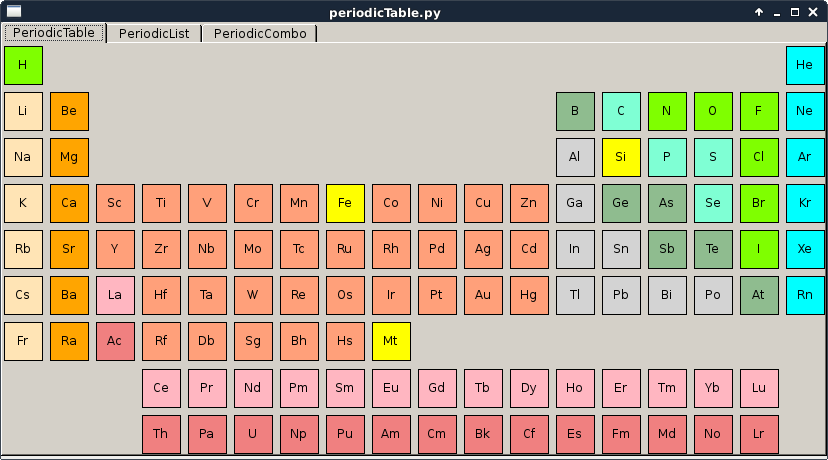
periodicTable.py |
|
This script is a simple example of how to use the periodic table widgets, select elements and connect signals. |
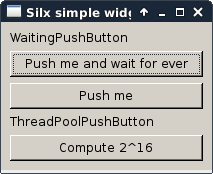
simplewidget.py |
|
This script shows a gallery of simple widgets provided by silx. It shows the following widgets:
|
viewer3DVolume.py |
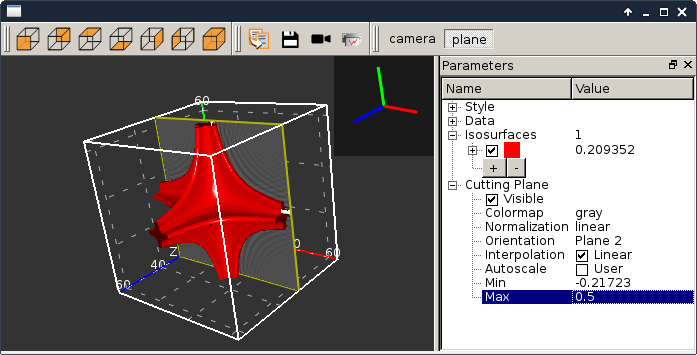
|
This script illustrates the use of silx.gui.plot3d.ScalarFieldView. It loads a 3D scalar data set from a file and displays iso-surfaces and an interactive cutting plane. It can also be started without providing a file. |
silx.gui.plot sample code¶
| Source | Screenshot | Description |
|---|---|---|
fftPlotAction.py,
fft.png |
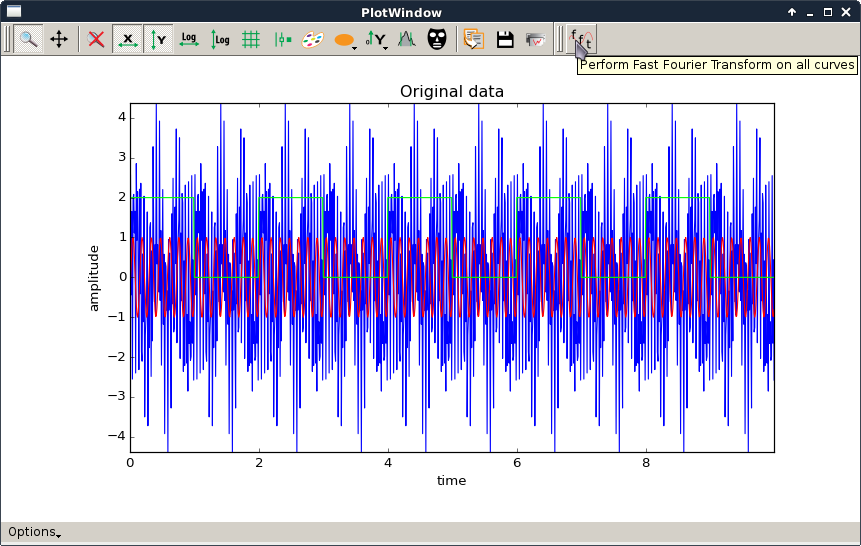
|
This script is a simple example of how to create a PlotWindow with a custom PlotAction added to the toolbar. The action computes the FFT of all curves and plots their amplitude spectrum. It also performs the reverse transform.
See shiftPlotAction.py for a simpler example with more basic comments. |
imageview.py |
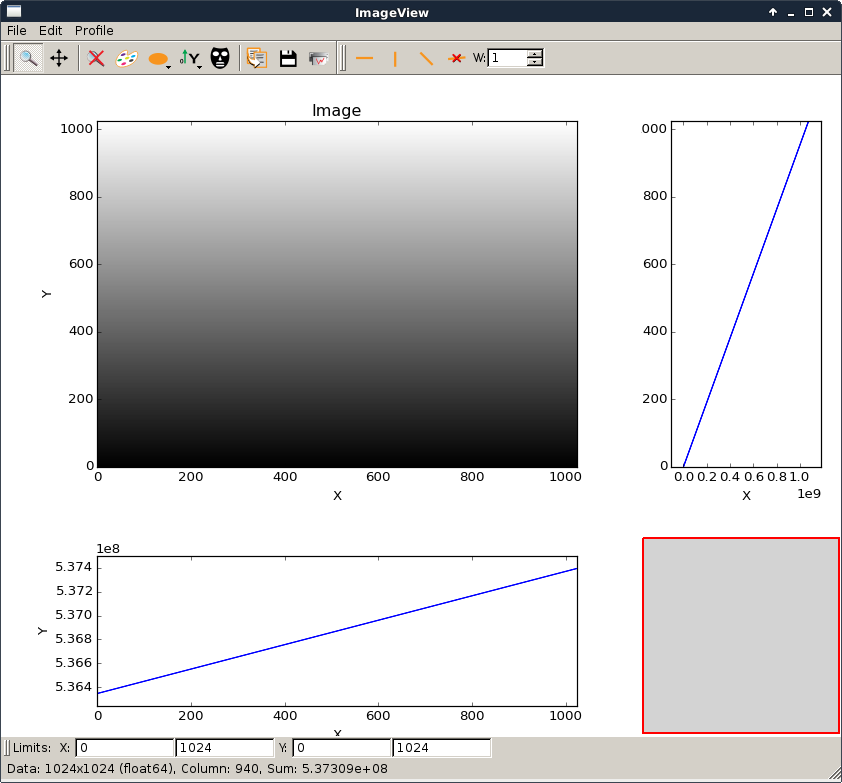
|
Example to show the use of ImageView widget. It can be used to open an EDF or TIFF file from the shell command line. To view an image file with the current installed silx library:
For developers with a git clone you can use it with the bootstrap To view an image file with the current installed silx library:
|
plotContextMenu.py |
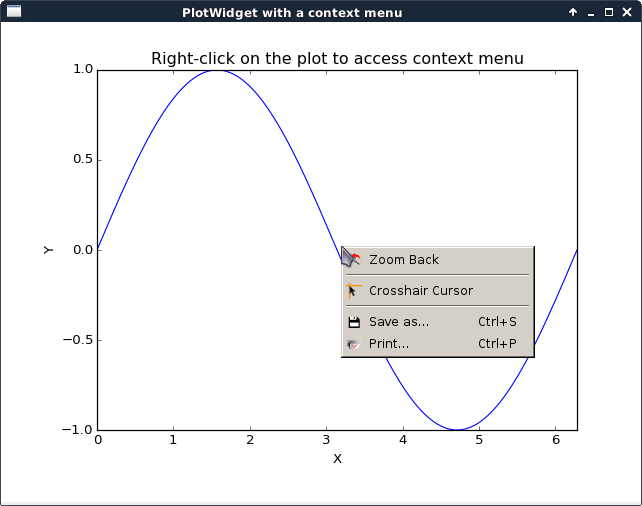
|
This script illustrates the addition of a context menu to a PlotWidget. This is done by adding a custom context menu to the plot area of PlotWidget: - set the context menu policy of the plot area to Qt.CustomContextMenu. - connect to the plot area customContextMenuRequested signal. The same method works with PlotWindow, Plot1D and Plot2D widgets as they inherit from PlotWidget. For more information on context menus, see Qt documentation. |
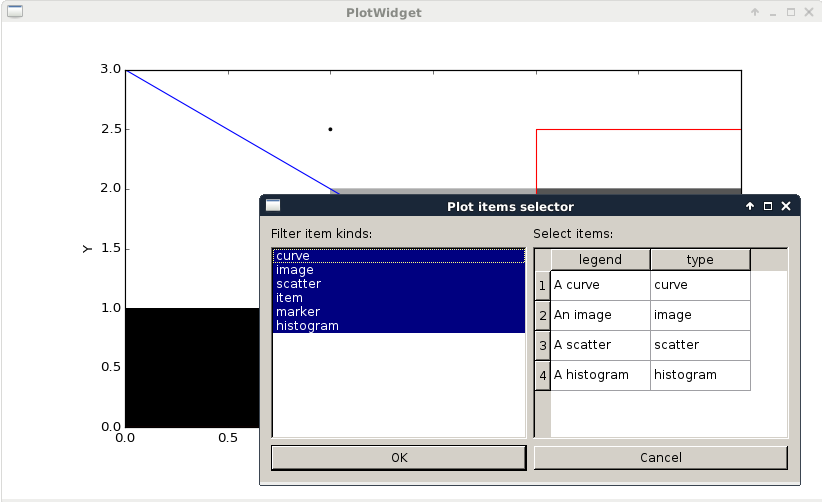
plotItemsSelector.py |
|
This example illustrates how to use a ItemsSelectionDialog widget
associated with a PlotWidget. |
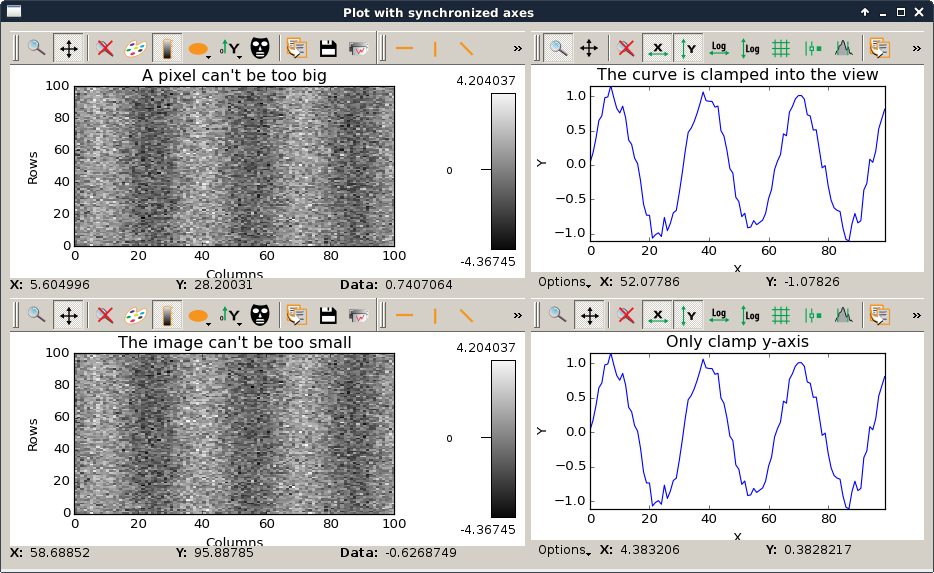
plotLimits.py |
|
This script is an example to illustrate how to use axis synchronization tool. |
plotUpdateFromThread.py |
|
This script illustrates the update of a silx.gui.plot widget from a thread. The problem is that plot and GUI methods should be called from the main thread. To safely update the plot from another thread, one need to make the update asynchronously from the main thread. In this example, this is achieved through a Qt signal. In this example we create a subclass of |
plotWidget.py |
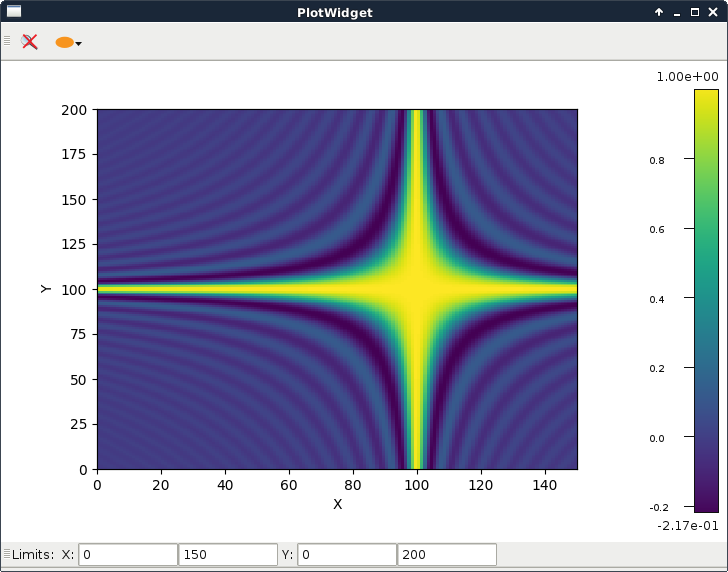
|
This script shows how to subclass It subclasses a
|
printPreview.py |
|
This script illustrates how to add a print preview tool button to any plot
widget inheriting Three plot widgets are instantiated. One of them uses a standalone
|
scatterMask.py |
|
This example demonstrates how to use ScatterMaskToolsWidget and NamedScatterAlphaSlider with a PlotWidget. |
shiftPlotAction.py |
|
This script is a simple (trivial) example of how to create a PlotWindow,
create a custom The action simply shifts the selected curve up by 1 unit by adding 1 to each value of y. |
stackView.py |
|
This script is a simple example to illustrate how to use the StackView widget. |
syncaxis.py |
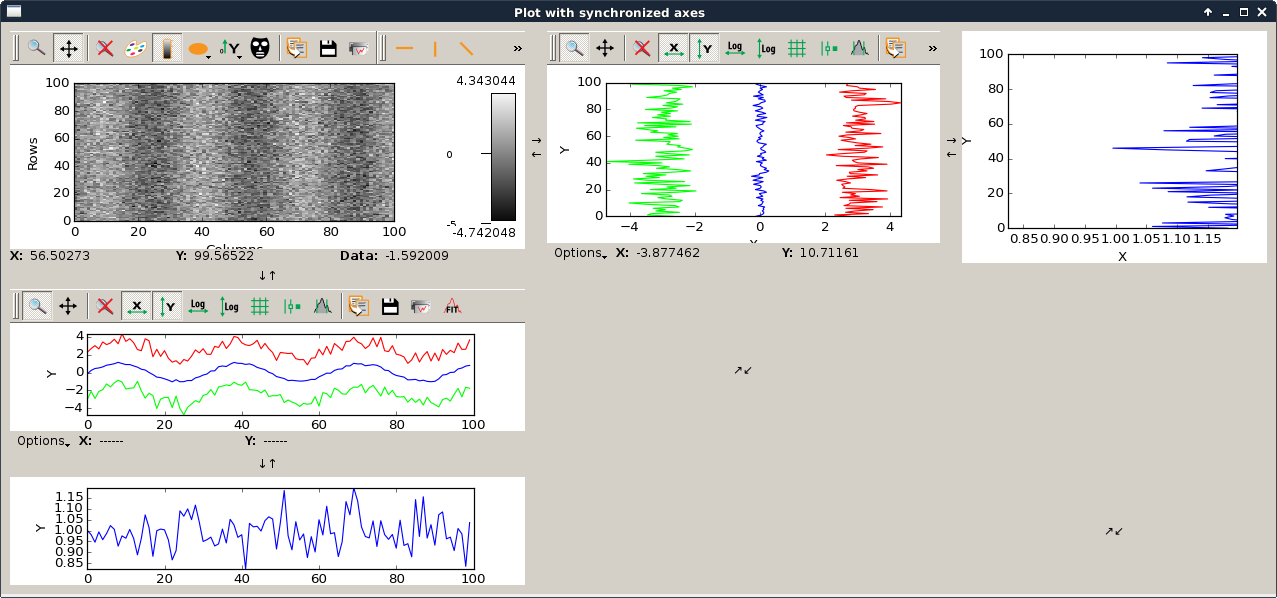
|
This script is an example to illustrate how to use axis synchronization tool. |
silx.io sample code¶
| Source | Screenshot | Description |
|---|---|---|
writetoh5.py |
This script converts a supported data file (SPEC, EDF...) to a HDF5 file. By default, it creates a new output file or fails if the output file given on the command line already exist, but the user can choose to overwrite an existing file, or append data to an existing HDF5 file. In case of appending data to HDF5 files, the user can choose between ignoring input data if a corresponding dataset already exists in the output file, or overwriting the existing dataset. By default, new scans are written to the root (/) of the HDF5 file, but it is possible to specify a different target path. |
