Integration tool: pyFAI-integrate¶
Purpose¶
PyFAI-integrate is a graphical interface (based on Python/Qt5) to perform azimuthal integration on a set of files. It exposes most of the important options available within pyFAI and allows you to select a GPU (or an openCL platform) to perform the calculation on.
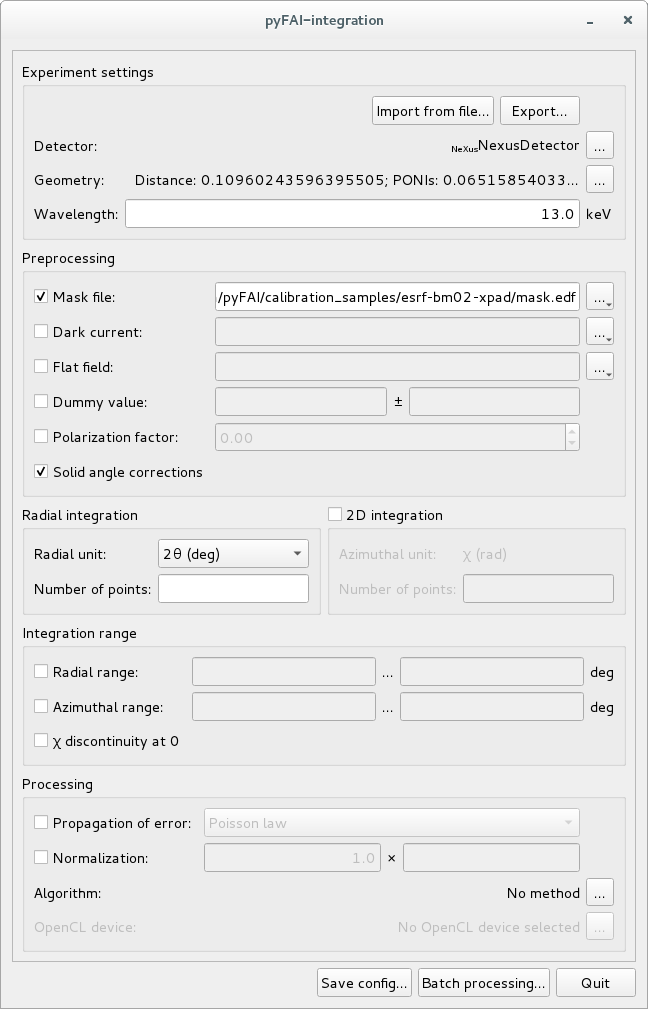
Usage¶
pyFAI-integrate [options] file1.edf file2.edf ...
Options:¶
- positional arguments:
FILE Files to be integrated
- optional arguments:
- -h, --help
show this help message and exit
- -V, --version
show program’s version number and exit
- -v, --verbose
switch to verbose/debug mode
- --debug
Set logging system in debug mode
- -o OUTPUT, --output OUTPUT
Directory or file where to store the output data
- -f FORMAT, --format FORMAT
output data format (can be HDF5)
- -s SLOW, --slow-motor SLOW
Dimension of the scan on the slow direction (makes sense only with HDF5)
- -r RAPID, --fast-motor RAPID
Dimension of the scan on the fast direction (makes sense only with HDF5)
- --no-gui
Process the dataset without showing the user interface.
- -j JSON, --json JSON
Configuration file containing the processing to be done
- --monitor-name MONITOR_KEY
Name of the monitor in the header of each input files. If defined the contribution of each input file is divided by the monitor. If the header does not contain or contains a wrong value, the contribution of the input file is ignored. On EDF files, values from ‘counter_pos’ can be accessed by using the expected mnemonic. For example ‘counter/bmon’.
- --delete
Delete the destination file if already exists
- --append
Append the processing to the destination file using an available group (HDF5 output)
- --overwrite
Overwrite the entry of the destination file if it already exists (HDF5 output)
Tips & Tricks:¶
PyFAI-integrate saves all parameters in a .azimint.json (hidden) file. This JSON file is an ascii file which can be edited and used to configure online data analysis using the LImA plugin of pyFAI.