Geometries in pyFAI¶
This notebook demonstrates the different orientations of axes in the geometry used by pyFAI.
Demonstration¶
The tutorial uses the Jypyter notebook.
In [1]:
import time
start_time = time.time()
%pylab nbagg
Populating the interactive namespace from numpy and matplotlib
In [2]:
import pyFAI
print("Using pyFAI version", pyFAI.version)
from pyFAI.gui import jupyter
from pyFAI.calibrant import get_calibrant
Using pyFAI version 0.15.0-beta2
/mntdirect/_scisoft/users/jupyter/jupy35/lib/python3.5/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.
from ._conv import register_converters as _register_converters
We will use a fake detector of 1000x1000 pixels of 100_µm each. The simulated beam has a wavelength of 0.1_nm and the calibrant chose is silver behenate which gives regularly spaced rings. The detector will originally be placed at 1_m from the sample.
In [3]:
wl = 1e-10
cal = get_calibrant("AgBh")
cal.wavelength=wl
detector = pyFAI.detectors.Detector(100e-6, 100e-6)
detector.max_shape=(1000,1000)
ai = pyFAI.AzimuthalIntegrator(dist=1, detector=detector)
ai.wavelength = wl
In [4]:
img = cal.fake_calibration_image(ai)
jupyter.display(img, label="Inital")
Out[4]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0dc9b128>
Translation orthogonal to the beam: poni1 and poni2¶
We will now set the first dimension (vertical) offset to the center of the detector: 100e-6 * 1000 / 2
In [5]:
p1 = 100e-6 * 1000 / 2
print("poni1:", p1)
ai.poni1 = p1
img = cal.fake_calibration_image(ai)
jupyter.display(img, label="set poni1")
poni1: 0.05
Out[5]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0dc5cdd8>
Let’s do the same in the second dimensions: along the horizontal axis
In [6]:
p2 = 100e-6 * 1000 / 2
print("poni2:", p2)
ai.poni2 = p2
print(ai)
img = cal.fake_calibration_image(ai)
jupyter.display(img, label="set poni2")
poni2: 0.05
Detector Detector Spline= None PixelSize= 1.000e-04, 1.000e-04 m
Wavelength= 1.000000e-10m
SampleDetDist= 1.000000e+00m PONI= 5.000000e-02, 5.000000e-02m rot1=0.000000 rot2= 0.000000 rot3= 0.000000 rad
DirectBeamDist= 1000.000mm Center: x=500.000, y=500.000 pix Tilt=0.000 deg tiltPlanRotation= 0.000 deg
Out[6]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0dc8f898>
The image is now properly centered. Let’s investigate the sample-detector distance dimension.
For this we need to describe a detector which has a third dimension which will be offseted in the third dimension by half a meter.
In [7]:
#define 3 plots
fig, ax = subplots(1, 3, figsize=(12,4))
import copy
ref_10 = cal.fake_calibration_image(ai, W=1e-4)
jupyter.display(ref_10, label="dist=1.0m", ax=ax[1])
ai05 = copy.copy(ai)
ai05.dist = 0.5
ref_05 = cal.fake_calibration_image(ai05, W=1e-4)
jupyter.display(ref_05, label="dist=0.5m", ax=ax[0])
ai15 = copy.copy(ai)
ai15.dist = 1.5
ref_15 = cal.fake_calibration_image(ai15, W=1e-4)
jupyter.display(ref_15, label="dist=1.5m", ax=ax[2])
Out[7]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0dc0ada0>
We test now if the sensot of the detector is not located at Z=0 in the detector referential but any arbitrary value:
In [8]:
class ShiftedDetector(pyFAI.detectors.Detector):
IS_FLAT = False # this detector is flat
IS_CONTIGUOUS = True # No gaps: all pixels are adjacents, speeds-up calculation
API_VERSION = "1.0"
aliases = ["ShiftedDetector"]
MAX_SHAPE=1000,1000
def __init__(self, pixel1=100e-6, pixel2=100e-6, offset=0):
pyFAI.detectors.Detector.__init__(self, pixel1=pixel1, pixel2=pixel2)
self.d3_offset = offset
def calc_cartesian_positions(self, d1=None, d2=None, center=True, use_cython=True):
res = pyFAI.detectors.Detector.calc_cartesian_positions(self, d1=d1, d2=d2, center=center, use_cython=use_cython)
return res[0], res[1], numpy.ones_like(res[1])*self.d3_offset
#This creates a detector offseted by half a meter !
shiftdet = ShiftedDetector(offset=0.5)
print(shiftdet)
Detector ShiftedDetector Spline= None PixelSize= 1.000e-04, 1.000e-04 m
In [9]:
aish = pyFAI.AzimuthalIntegrator(dist=1, poni1=p1, poni2=p2, detector=shiftdet, wavelength=wl)
print(aish)
shifted = cal.fake_calibration_image(aish, W=1e-4)
jupyter.display(shifted, label="dist=1.0m, offset Z=+0.5m")
Detector ShiftedDetector Spline= None PixelSize= 1.000e-04, 1.000e-04 m
Wavelength= 1.000000e-10m
SampleDetDist= 1.000000e+00m PONI= 5.000000e-02, 5.000000e-02m rot1=0.000000 rot2= 0.000000 rot3= 0.000000 rad
DirectBeamDist= 1000.000mm Center: x=500.000, y=500.000 pix Tilt=0.000 deg tiltPlanRotation= 0.000 deg
Out[9]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0dc8f358>
This image is the same as the one with dist=1.5m The positive distance along the d3 direction is equivalent to increase the distance. d3 is in the same direction as the incoming beam.
After investigation of the three translations, we will now investigate the rotation along the different axes.
Investigation on the rotations:¶
Any rotations of the detector apply after the 3 translations (dist, poni1 and poni2)
The first axis is the vertical one and a rotation around it ellongates ellipses along the orthogonal axis:
In [10]:
rotation = +0.2
ai.rot1 = rotation
print(ai)
img = cal.fake_calibration_image(ai)
jupyter.display(img, label="rot1 = 0.2 rad")
Detector Detector Spline= None PixelSize= 1.000e-04, 1.000e-04 m
Wavelength= 1.000000e-10m
SampleDetDist= 1.000000e+00m PONI= 5.000000e-02, 5.000000e-02m rot1=0.200000 rot2= 0.000000 rot3= 0.000000 rad
DirectBeamDist= 1020.339mm Center: x=-1527.100, y=500.000 pix Tilt=11.459 deg tiltPlanRotation= 180.000 deg
Out[10]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0dbb44e0>
So a positive rot1 is equivalent to turning the detector to the right, around the sample position (where the observer is).
Let’s consider now the rotation along the horizontal axis, rot2:
In [11]:
rotation = +0.2
ai.rot1 = 0
ai.rot2 = rotation
print(ai)
img = cal.fake_calibration_image(ai)
jupyter.display(img, label="rot2 = 0.2 rad")
Detector Detector Spline= None PixelSize= 1.000e-04, 1.000e-04 m
Wavelength= 1.000000e-10m
SampleDetDist= 1.000000e+00m PONI= 5.000000e-02, 5.000000e-02m rot1=0.000000 rot2= 0.200000 rot3= 0.000000 rad
DirectBeamDist= 1020.339mm Center: x=500.000, y=2527.100 pix Tilt=11.459 deg tiltPlanRotation= 90.000 deg
Out[11]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0db81f28>
In [12]:
rotation = +0.2
ai.rot1 = rotation
ai.rot2 = rotation
ai.rot3 = 0
print(ai)
img = cal.fake_calibration_image(ai)
jupyter.display(img, label="rot1 = rot2 = 0.2 rad")
Detector Detector Spline= None PixelSize= 1.000e-04, 1.000e-04 m
Wavelength= 1.000000e-10m
SampleDetDist= 1.000000e+00m PONI= 5.000000e-02, 5.000000e-02m rot1=0.200000 rot2= 0.200000 rot3= 0.000000 rad
DirectBeamDist= 1041.091mm Center: x=-1527.100, y=2568.329 pix Tilt=16.151 deg tiltPlanRotation= 134.423 deg
Out[12]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0db2a9b0>
In [13]:
rotation = +0.2
import copy
ai2 = copy.copy(ai)
ai2.rot1 = rotation
ai2.rot2 = rotation
ai2.rot3 = rotation
print(ai2)
img2 = cal.fake_calibration_image(ai2)
jupyter.display(img2, label="rot1 = rot2 = rot3 = 0.2 rad")
Detector Detector Spline= None PixelSize= 1.000e-04, 1.000e-04 m
Wavelength= 1.000000e-10m
SampleDetDist= 1.000000e+00m PONI= 5.000000e-02, 5.000000e-02m rot1=0.200000 rot2= 0.200000 rot3= 0.200000 rad
DirectBeamDist= 1041.091mm Center: x=-1527.100, y=2568.329 pix Tilt=16.151 deg tiltPlanRotation= 134.423 deg
Out[13]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0dadb630>
If one considers the rotation along the incident beam, there is no visible effect on the image as the image is invariant along this transformation.
To actually see the effect of this third rotation one needs to perform the azimuthal integration and display the result with properly labeled axes.
In [14]:
fig, ax = subplots(1,2,figsize=(10,5))
res1 = ai.integrate2d(img, 300, 360, unit="2th_deg")
jupyter.plot2d(res1, label="rot3 = 0 rad", ax=ax[0])
res2 = ai2.integrate2d(img2, 300, 360, unit="2th_deg")
jupyter.plot2d(res2, label="rot3 = 0.2 rad", ax=ax[1])
Out[14]:
<matplotlib.axes._subplots.AxesSubplot at 0x7fdd0daa6be0>
So the increasing rot3 creates more negative azimuthal angles: it is like rotating the detector clockwise around the incident beam.
Conclusion¶
All 3 translations and all 3 rotations can be summarized in the following figure:
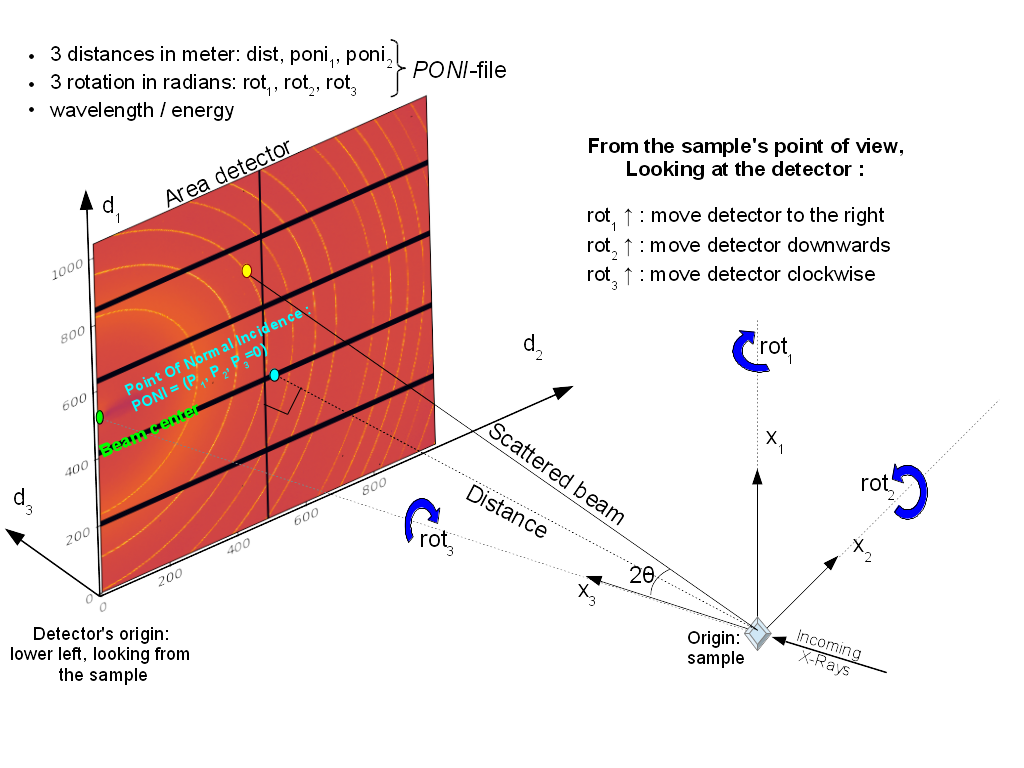
PONI figure
It may appear strange to have (x_1, x_2, x_3) indirect indirect but this has been made in such a way chi, the azimuthal angle, is 0 along x_2 and 90_deg along x_1 (and not vice-versa).
In [15]:
print("Processing time: %.3fs"%(time.time()-start_time))
Processing time: 3.287s