Integration tool: diff_map¶
Purpose¶
Azimuthal integration for diffraction imaging
Diffraction mapping is an imaging experiment where 2D diffraction patterns are recorded while performing a 2D scan along two axes, one slower and the other fastest.
Diff_map provides a Graphical User Interface (based on top of PyQt, pyFAI and h5py) which allows the reduction of this 4D dataset into a 3D dataset containing the two dimension of movement and the many diffraction angles (the output can be q-space for PDF-CT).
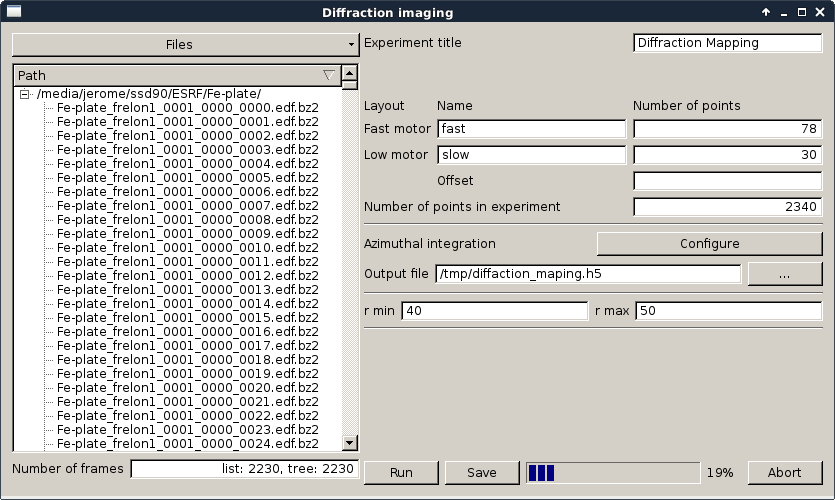
On the left-hand side, the user can select a bunch of files to be processed. For now, any image format supported by FabIO is possible, including multi-frame images, but not ye NeXus files (work ongoing).
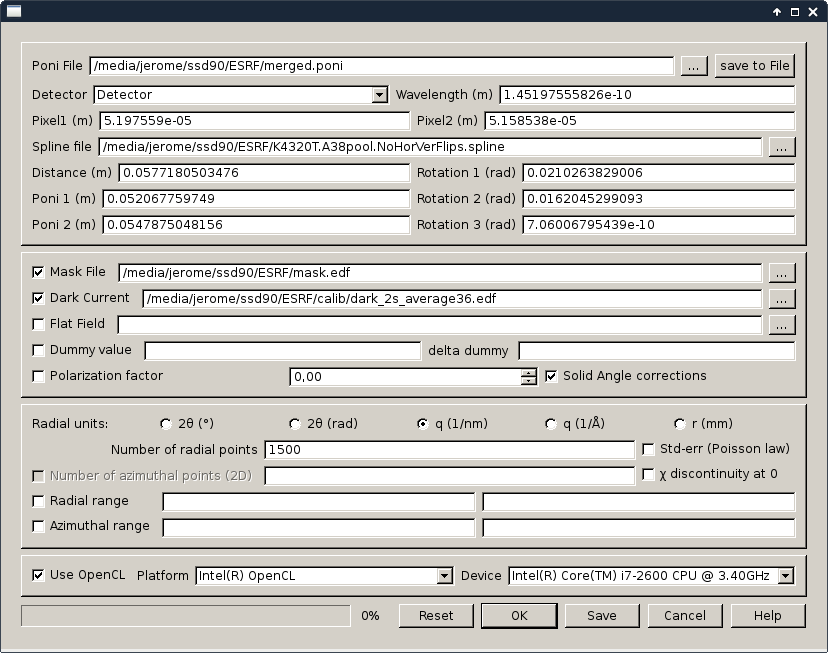
On the right-hand side, the motor range can be specified togeather with their names. The diffraction parameters can be setup in a similar way to pyFAI-integrate. The output name can also be specified.
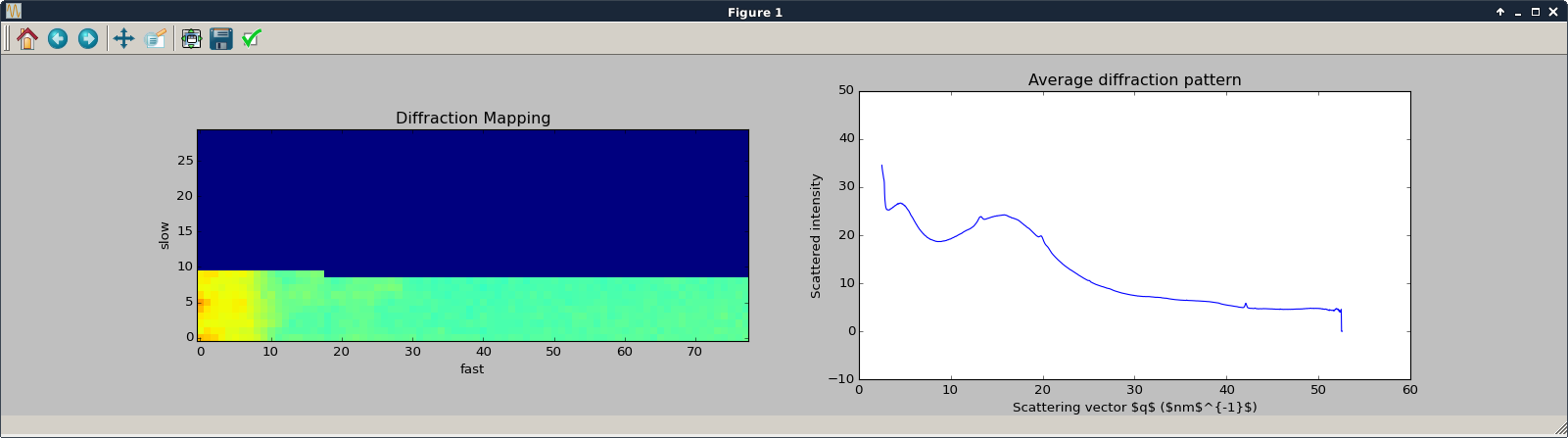
The processing is launched using the Run button which opens a matplotlib figure to display the plot of the diffraction pattern and of the map image under construction. During this processing, one can select a scatering range to highlight the regions of interest.
Further analysis can be performed on the resulting dataset using the PyMca roitool where the 1d dataset has to be selected as last dimension. The result file aims at NeXus compliance.
This tool can be used for tomography experiments if one considers the slow scan direction as the rotation.
The user interface can be disabled to allow scripting interface and providing a JSON configuration file (or all the options).
Usage:¶
diff_map [options] imagefiles*
- positional arguments:
- FILE List of files to integrate
- optional arguments:
-h, --help show this help message and exit -V, --version show program’s version number and exit -o FILE, --output FILE HDF5 File where processed map will be saved -v, --verbose switch to verbose/debug mode, defaut: quiet -P FILE, --prefix FILE Prefix or common base for all files -e EXTENSION, --extension EXTENSION Process all files with this extension -t FAST, --fast FAST number of points for the fast motion. Mandatory without GUI -r SLOW, --slow SLOW number of points for slow motion. Mandatory without GUI -c NPT_RAD, --npt NPT_RAD number of points in diffraction powder pattern, Mandatory without GUI -d FILE, --dark FILE list of dark images to average and subtract -f FILE, --flat FILE list of flat images to average and divide -m FILE, --mask FILE file containing the mask, no mask by default -p FILE, --poni FILE file containing the diffraction parameter (poni-file), Mandatory without GUI -O OFFSET, --offset OFFSET do not process the first files -g, --gpu process using OpenCL on GPU -S, --stats show statistics at the end --gui Use the Graphical User Interface --no-gui Do not use the Graphical User Interface --config CONFIG provide a JSON configuration file
Bugs:¶
- If the number of files is too large, use double quotes “*.edf”
- There is a known bug on Debian7 where importing a large number of file can take much longer than the integration itself: consider passing files in the command line
$ diff_map --help
usage: diff_map [options] -p ponifile imagefiles*
If the number of files is too large, use double quotes like "*.edf"
Azimuthal integration for diffraction imaging. Diffraction mapping is an
experiment where 2D diffraction patterns are recorded while performing a 2D
scan. Diff_map is a graphical application (based on pyFAI and h5py) which
allows the reduction of this 4D dataset into a 3D dataset containing the two
motion dimensions and the many diffraction angles (thousands). The resulting
dataset can be opened using PyMca roitool where the 1d dataset has to be
selected as last dimension. This result file aims at being NeXus compliant.
This tool can be used for diffraction tomography experiment as well,
considering the slow scan direction as the rotation.
positional arguments:
FILE List of files to integrate. Mandatory without GUI
optional arguments:
-h, --help show this help message and exit
-V, --version show program's version number and exit
-o FILE, --output FILE
HDF5 File where processed map will be saved. Mandatory
without GUI
-v, --verbose switch to verbose/debug mode, default: quiet
-P FILE, --prefix FILE
Prefix or common base for all files
-e EXTENSION, --extension EXTENSION
Process all files with this extension
-t FAST, --fast FAST number of points for the fast motion. Mandatory
without GUI
-r SLOW, --slow SLOW number of points for slow motion. Mandatory without
GUI
-c NPT_RAD, --npt NPT_RAD
number of points in diffraction powder pattern.
Mandatory without GUI
-d FILE, --dark FILE list of dark images to average and subtract (comma
separated list)
-f FILE, --flat FILE list of flat images to average and divide (comma
separated list)
-m FILE, --mask FILE file containing the mask, no mask by default
-p FILE, --poni FILE file containing the diffraction parameter (poni-file),
Mandatory without GUI
-O OFFSET, --offset OFFSET
do not process the first files
-g, --gpu process using OpenCL on GPU
-S, --stats show statistics at the end
--gui Use the Graphical User Interface
--no-gui Do not use the Graphical User Interface
--config CONFIG provide a JSON configuration file
Bugs: Many, see hereafter: 1)If the number of files is too large, use double
quotes "*.edf" 2)There is a known bug on Debian7 where importing a large
number of file can take much longer than the integration itself: consider
passing files in the command line